Set Up¶
Choose computing environment¶
There are two ways to follow this tutorial:
A) Use the pangeo binder. This is a good option if you don't have access to a Unix/Linux computer.
-
Open the binder environment in a new tab (i.e., by typing Ctrl and clicking link). It will take a few minutes to build:
-
Click on the "Terminal" button to launch it:
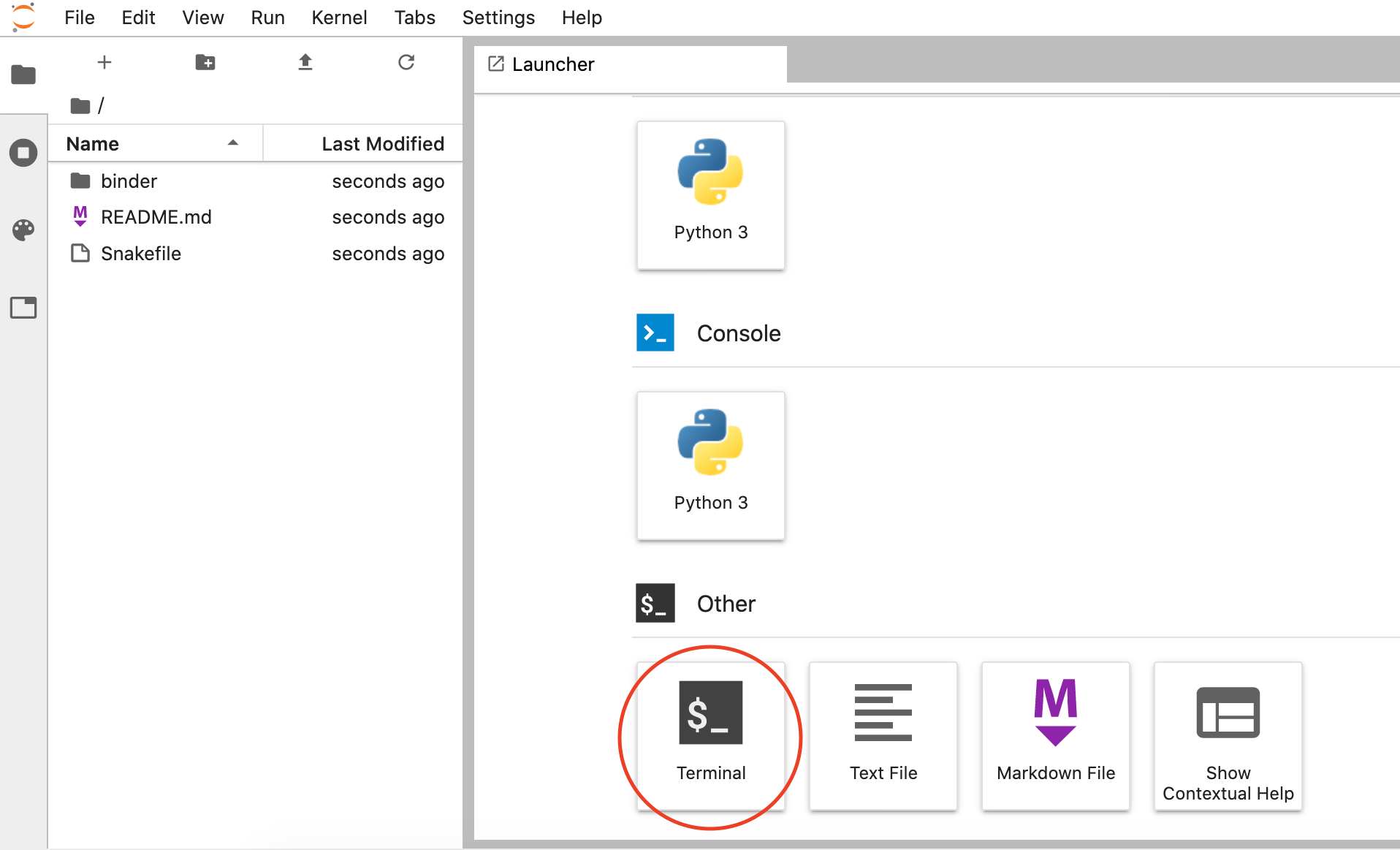
-
Then, follow the set up instructions below starting at step 2. The Snakefile and necessary conda environment files are already installed in the binder.
Warning
When you close the binder, it does NOT save your work so download any files you want to keep.
-
To download files, right-click the file you want to save, and select "Download":
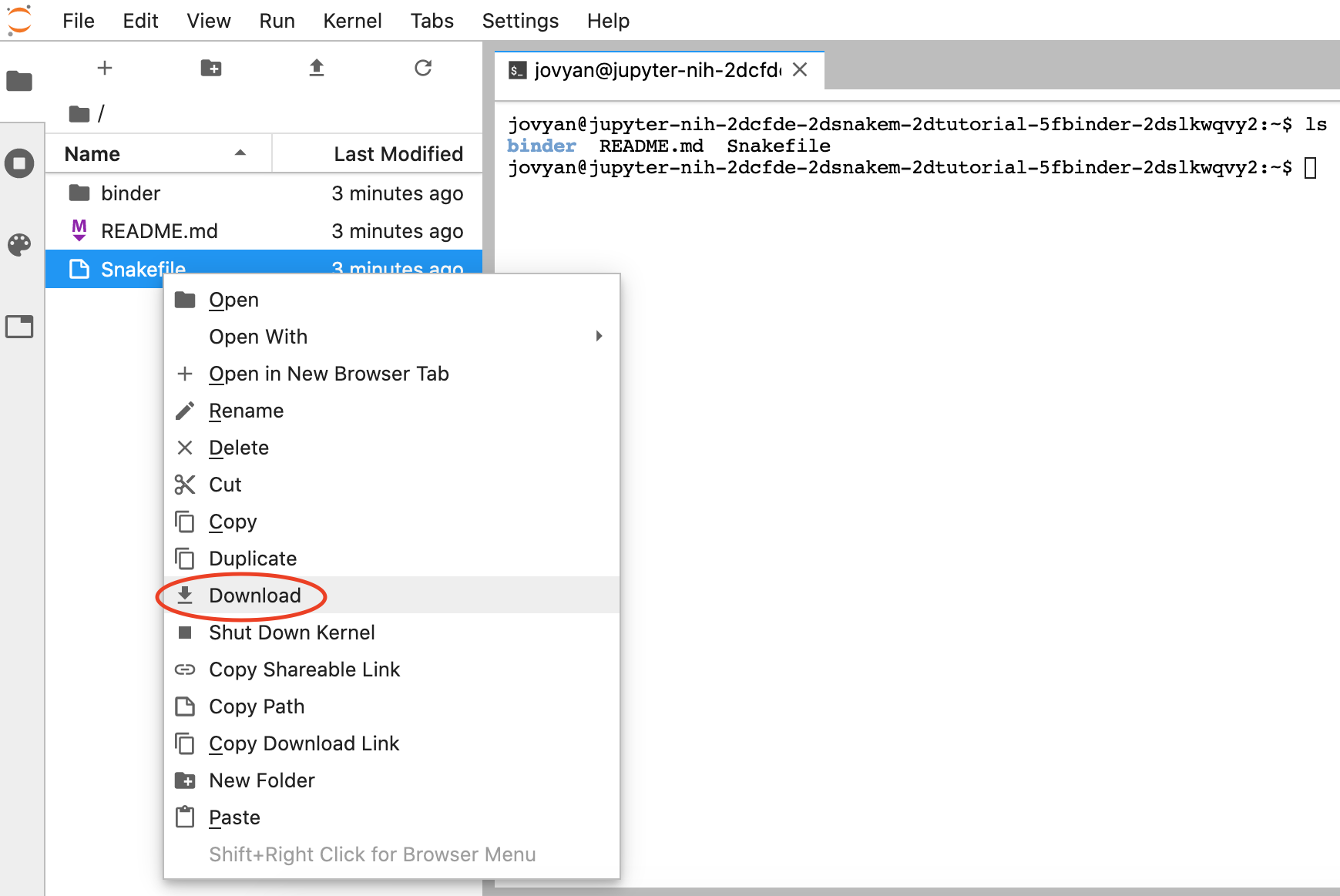
-
To close the binder, go to "File" and click "Shut Down":
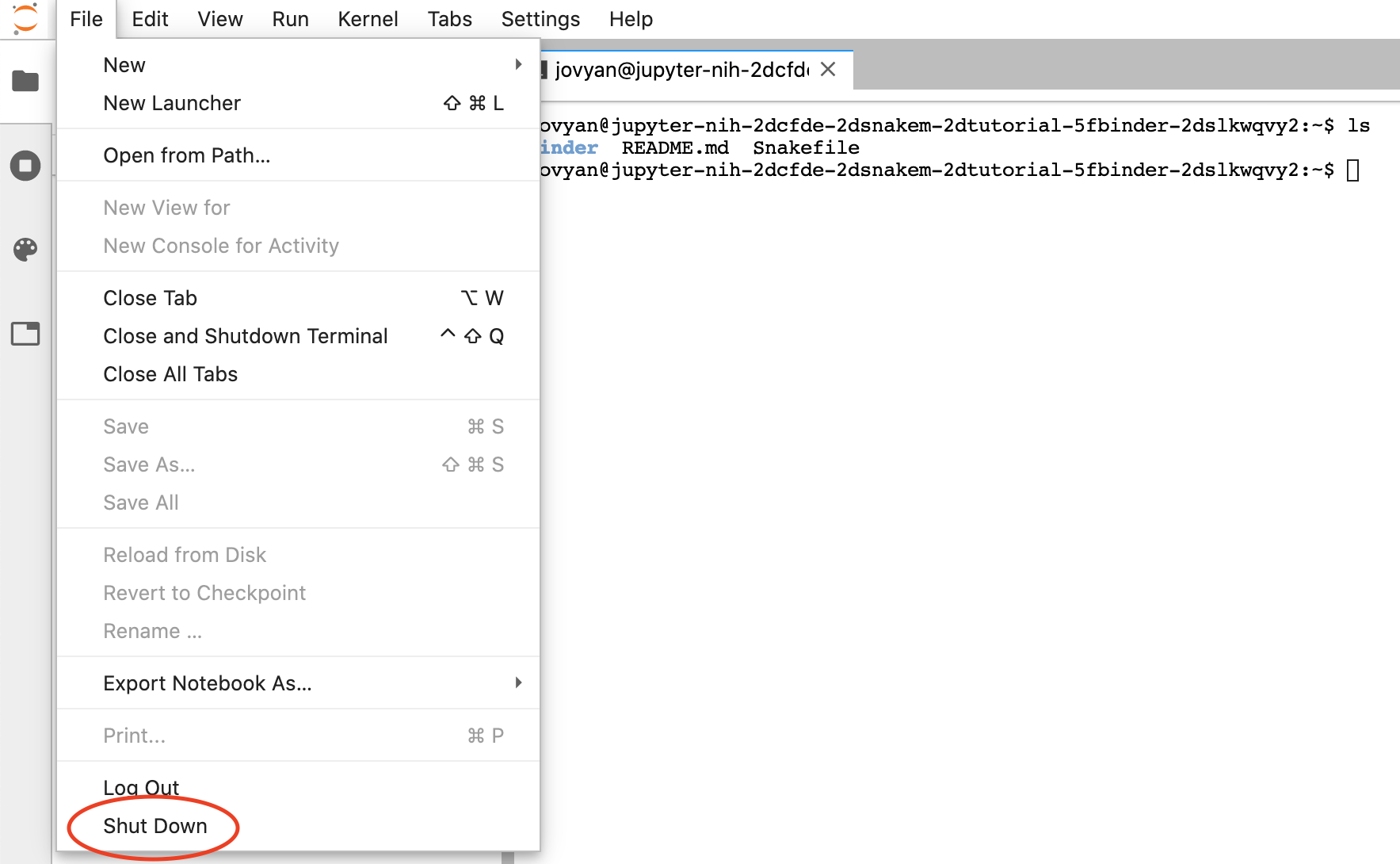
Note
We also have an Rstudio version of the binder environment. The only difference compared to the terminal binder environment is the layout of Rstudio panels. The template Snakefile and software are pre-loaded with the binder.
B) Use a Unix or Linux environment (e.g., with a Mac or High Performing Computing environment).
- For this option, please follow all the set up instructions below.
Set up computing environment¶
We will use conda to create an computer software for this tutorial. If you don't have conda installed, please see the Set up computing environment with conda on MacOS tutorial.
Tip
Please refer to the conda command cheat sheet for commonly used conda commands!
Step 1: Download tutorial files:¶
First, create a new directory for this tutorial in the (base) conda environment, e.g.,:
mkdir learn_snakemake
We need two files for this tutorial. Click the links and save them in the directory you created above: 1) environment.yml and 2) Snakefile.
Rename the "Snakefile.py" to "Snakefile". There should be no file extension (we just added it so you'd be able to download the file!).
If you want to use wget to download them to a remote computer, these commands should work on most Linux systems:
wget https://training.nih-cfde.org/en/latest/General-Tools/Snakemake/snakemake_tutorial_docs/environment.yml
wget https://training.nih-cfde.org/en/latest/General-Tools/Snakemake/snakemake_tutorial_docs/Snakefile.py
mv Snakefile.py Snakefile
Step 2: Create new conda environment:¶
conda is a software installation system that we will not cover in any detail today, but it is how we install all of our software for this lesson! Please see our Introduction to Conda for more information.
The environment.yml file tells conda 1) where to look for the software installations under "channels" and 2) what software to install under "dependencies". You can also specify specific software versions, otherwise conda will download the most up-to-date version. Here are the specifications we'll use for this tutorial, as described in environment.yml:
channels (where the software will be installed from):
- conda-forge
- bioconda
- defaults
dependencies (which software will be installed):
- bwa
- snakemake-minimal=6.3.0
- samtools=1.10
- bcftools
If you're using the pangeo binder, the steps below are already done for you; if you are on a laptop or other computer, you can install all the necessary software by running the following command:
conda env create -n snaketest -f environment.yml
This creates a new environment called snaketest, which you can then activate.
Step 3: Activate conda environment:¶
To use the software you just installed, execute:
conda activate snaketest
Your terminal command prompt should now look like (snaketest) $ instead of (base) $. This indicates that we can use all of the software installed by conda for this lesson.
Step 4: Test that your environment is ready to go¶
You should have several software packages installed in your snaketest environment now. Check it out!
samtools --version
samtools 1.10
Using htslib 1.10.2
Copyright (C) 2019 Genome Research Ltd.
If you get an error, the software installation may have failed. You can check the software that is installed in your conda environment: conda list
To leave the conda environment, type: conda deactivate
Later in the tutorial, we'll use wget to download data. Installing wget on MacOS can be achieved with conda install. This step will take a few minutes and the installation should be done in the base conda environment:
Go back to base environment:
conda deactivate
Install wget:
conda install -c conda-forge wget
Test installation:
wget --version
GNU Wget 1.20.1 built on darwin14.5.0.
-cares +digest -gpgme +https +ipv6 -iri +large-file -metalink -nls
+ntlm +opie -psl +ssl/openssl
Wgetrc:
/Users/amanda/miniconda3/envs/snaketest/etc/wgetrc (system)
Compile:
x86_64-apple-darwin13.4.0-clang -DHAVE_CONFIG_H
-DSYSTEM_WGETRC="/Users/amanda/miniconda3/envs/snaketest/etc/wgetrc"
-DLOCALEDIR="/Users/amanda/miniconda3/envs/snaketest/share/locale"
-I. -I../lib -I../lib -D_FORTIFY_SOURCE=2 -mmacosx-version-min=10.9
-I/Users/amanda/miniconda3/envs/snaketest/include -DHAVE_LIBSSL
-I/Users/amanda/miniconda3/envs/snaketest/include -DNDEBUG
-march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE
-fstack-protector-strong -O2 -pipe
-I/Users/amanda/miniconda3/envs/snaketest/include
-fdebug-prefix-map=/opt/concourse/worker/volumes/live/c2196a4d-0070-4d98-71cc-9baa5fb53ac9/volume/wget_1551978005217/ work=/usr/local/src/conda/wget-1.20.1
-fdebug-prefix-map=/Users/amanda/miniconda3/envs/snaketest=/usr/local/src/conda-prefix
Link:
x86_64-apple-darwin13.4.0-clang
-I/Users/amanda/miniconda3/envs/snaketest/include -DHAVE_LIBSSL
-I/Users/amanda/miniconda3/envs/snaketest/include -DNDEBUG
-march=core2 -mtune=haswell -mssse3 -ftree-vectorize -fPIC -fPIE
-fstack-protector-strong -O2 -pipe
-I/Users/amanda/miniconda3/envs/snaketest/include
-fdebug-prefix-map=/opt/concourse/worker/volumes/live/c2196a4d-0070-4d98-71cc-9baa5fb53ac9/volume/wget_1551978005217/work=/usr/local/src/conda/wget-1.20.1
-fdebug-prefix-map=/Users/amanda/miniconda3/envs/snaketest=/usr/local/src/conda-prefix
-Wl,-pie -Wl,-headerpad_max_install_names -Wl,-dead_strip_dylibs
-Wl,-rpath,/Users/amanda/miniconda3/envs/snaketest/lib
-L/Users/amanda/miniconda3/envs/snaketest/lib
-L/Users/amanda/miniconda3/envs/snaketest/lib -lssl -lcrypto
-L/Users/amanda/miniconda3/envs/snaketest/lib -lz ftp-opie.o
openssl.o http-ntlm.o ../lib/libgnu.a -lcrypto
Copyright (C) 2015 Free Software Foundation, Inc.
License GPLv3+: GNU GPL version 3 or later
<http://www.gnu.org/licenses/gpl.html>.
This is free software: you are free to change and redistribute it.
There is NO WARRANTY, to the extent permitted by law.
Originally written by Hrvoje Niksic <[email protected]>.
Please send bug reports and questions to <[email protected]>.